| Index to this page |
Histocompatibility molecules are glycoproteins expressed at the surface of almost all vertebrate cells. They get their name because they are responsible for the compatibility — or rather the lack of it — of the tissues of genetically different individuals. Monozygotic ("identical") human twins have the same histocompatibility molecules on their cells, and they can accept transplants of tissue from each other. The rest of us have a set of histocompatibility molecules that is probably unique to us. A graft of our tissue into another human will provoke an immune response which, if left unchecked, will end in the rejection of the transplant. So the histocompatibility molecules of one individual act as antigens when introduced into a different individual. In fact, the histocompatibility molecules are often called histocompatibility antigens or transplantation antigens.
| Link to discussion of transplant rejection. |
The most rapid and severe rejection of foreign tissue occurs when there is a failure to properly match the donor and recipient for the major histocompatibility molecules. There are two categories: class I and class II.

Class I molecules consist of two polypeptide chains, a long one (on the left) of 346 amino acids — it is called the heavy chain — and a short one (on the right) of 99 amino acids.
The heavy chain consists of 5 main regions or domains:To the right is the protein molecule called beta-2 microglobulin ("β2M"). It is not attached to the heavy chain by any covalent bonds, but rather by a number of noncovalent interactions.
The dark bars represent disulfide (S-S) bridges linking portions of each domain (except the N domain). However, the bonds in S-S bridges are no longer than any other covalent bond, so if this molecule could be viewed in its actual tertiary (3D) configuration, we would find that the portions of the polypeptide chains containing the linked Cys are actually close together.
The outermost domains ("N" and "C1") contain two segments of alpha helix that form two ridges with a groove between them. A small molecule (e.g., a short peptide) is attached noncovalently in the groove between the two alpha helices, rather like a hot dog in a bun (not shown here, but look below).
| You can also link to a color model (92K) showing these features. But note that the terminology for the domains is different in this model: N = α1, C1 = α2, C2 = α 3 |
The two objects on the left of the image that look like candelabra represent the short, branched chains of sugars in this glycoprotein.
The regions marked "Papain" represent the places on the heavy chain that are attacked by the proteinase papain (and made it possible to release the extracellular domains from the plasma membrane for easier analysis).
This image (courtesy of T.J. Kindt and J. E. Coligan) represents the structure of a class I histocompatibility molecule, called H-2K. Almost all the cells of an animal's body (in this case, a mouse) have thousands of these molecules present in their plasma membrane. These molecules provide tissue identity and serve as major targets in the rejection of transplanted tissue and organs. But tissue rejection is not their natural function. Class I molecules serve to display antigens on the surface of the cell so that they can be "recognized" by T cells.
Humans synthesize six different types of class I molecules. However, we will examine only the three "major" ones: HLA-A, HLA-B, and HLA-C. (HLA stands for human leukocyte antigen; because the molecules were first studied on leukocytes). These differ only in their heavy chain, all sharing the same type of beta-2 microglobulin. The genes encoding the different heavy chains are clustered on chromosome 6 in the major histocompatibility complex (MHC).
| Link to a discussion of the MHC of humans. |
We inherit a gene for each of the three types of heavy chain from each parent so it is possible, in fact common, to express two allelic versions of each type. Thus a person heterozygous for HLA-A, HLA-B, and HLA-C expresses six different class I proteins. These are synthesized and displayed by most of the cells of the body (except those of the central nervous system).

Although histocompatibility molecules were discovered because of the crucial role they play in graft rejection, clearly evolution did not give vertebrates these molecules for that function. So what is their normal function? The answer: to display antigens so that they can be "seen" by T lymphocytes.
The antigen receptor on T lymphocytes (or T cells, as they are commonly called) "sees" an epitope that is a mosaic of the small molecule in the groove and portions of the alpha helices flanking it.
| Discussion of antigen presentation to T cells. |
The small molecules ("hot dogs") are enormously diverse. They probably represent fragments derived from all the proteins present within the cell. These would include:
Human class II molecules are designated HLA-D, and the genes encoding them are also located in the major histocompatibility complex (MHC). Class II molecules consist of two transmembrane polypeptides. These interact to form a groove at their outer end which, like class I molecules, always contains a fragment of antigen. But the fragments bound to class II molecules are derived from antigens that the cell has taken in from its surroundings. Extracellular molecules are engulfed by endocytosis. The endosomes fuse with lysosomes and their contents are partially digested. The resulting fragments are placed in class II molecules and returned to the cell surface.
| See Antigen Presentation for more details |
Class II molecules, in contrast to class I, are normally expressed on only certain types of cells. These are cells like macrophages and B lymphocytes that specialize in processing and presenting extracellular antigens to T lymphocytes. Thus antigen presentation by class II molecules differs from that by class I in two important ways:
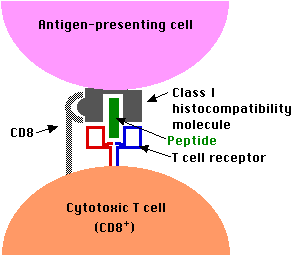
Because of the need for CD8 to bind to a receptor site found only on class I histocompatibility molecules, CD8+ T cells are only able to respond to antigens presented by class I molecules. Most CD8+ T cells are cytotoxic T cells (CTLs). They contain the machinery for destroying cells whose class I epitope they recognize.
An example:Every time you get a viral infection, say influenza (flu), the virus invades certain cells of your body. Once inside, the virus subverts the metabolism of the cell to make more virus. This involves synthesizing molecules encoded by the viral genome. In due course, these are assembled into a fresh crop of virus particles that leave the cell (often killing it in the process) and spread to new cells.
Except while in transit from their old home to their new, the virus works inside cells safe from any antibodies. But early in their intracellular life, infected cells display fragments of the viral proteins being synthesized in the cytoplasm in their surface class I molecules.
Any cytotoxic T cells specific for that antigen will bind to the infected cell and often will be able to destroy it before it can release a fresh crop of virus.
The bottom line: the function of the body's CD8+ T cells is to monitor all the cells of the body ready to destroy any that express foreign antigen fragments in their class I molecules.
| Further details of how infected cells present viral antigens to CD8+ T cells |
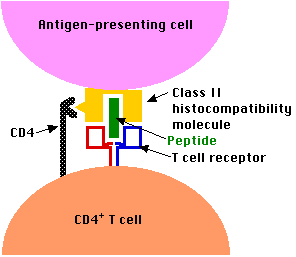 The CD4 molecules expressed on the surface of CD4+ T cells enable them to bind to cells presenting antigen fragments in class II molecules but not in class I. Only certain types of cells, those specialized for taking up antigen from extracellular fluids, express class II molecules. Among the most important of these are
The CD4 molecules expressed on the surface of CD4+ T cells enable them to bind to cells presenting antigen fragments in class II molecules but not in class I. Only certain types of cells, those specialized for taking up antigen from extracellular fluids, express class II molecules. Among the most important of these are
| Further discussion with graphic. |
Activated T cells
| Discussion of Helper T cells |
| Welcome&Next Search |