| Index to this page |
Advances in genetics — especially the sequencing of entire genomes from a wide variety of animals — has revealed an unexpected paradox.
While the animal kingdom contains an extraordinary diversity of body types:| (These links will remind you of the diversity of both invertebrates and vertebrates.) |
the great structural diversity of animals is not reflected in their genetic makeup.
Throughout the animal kingdom, one finds thousands of orthologous genes; that is, genes that have similar sequences [examples] and encode similar products [examples].
At the level of the cell, and even of tissues, this perhaps should not be surprising. After all, most types of cells — from whatever animal — are quite similar in their structure and function. Thus we would expect that their genes that encode ribosomal proteins, cytochromes, histones, etc., etc. would be similar.
In trying to resolve the paradox that these findings present, it is useful to distinguish betweenHousekeeping genes generally
Even such primitive animals as sponges and cnidarians have hundreds of toolkit genes that are clear orthologs to genes of humans.
Some examples are genes whose products are involved in cell signaling (e.g., Wnt and β-catenin, Hedgehog, Notch, Receptor Tyrosine Kinases (RTKs), components of JAK/STAT pathways, and Transforming Growth Factor-beta TGF-β receptors).
In fact the sequences of many of these genes from different animal phyla are so similar that they can be interchanged!
This similarity can be tested in animals that can be made transgenic. Some examples:A mutation that would be lethal in the protein coding region of a gene need not be if it occurs in a control region (e.g. promoters and/or enhancers) of that gene.
In fact, there is increasing evidence that mutations in control regions have played an important part in evolution. Examples:Pitx1 is a
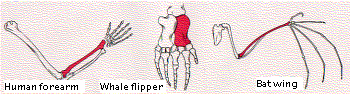
When we consider the dramatically-different activities that a given toolkit gene product can perform in different parts of the same animal, it is easier to understand how easy it must be for these same genes to alter the structure of the same body part in different species, e.g., the human arm and the wing of the bat.
Pitx1 is an essential gene. Mutations in the coding regions are lethal when homozygous (shown in mice).
However, mutations in noncoding regions need not be.
All vertebrates have a pelvic girdle with associated bones which make upPitx1 is needed by them all for the proper development of these structures (as well as the other functions of Pitx1).
In a remarkable study of three-spined sticklebacks published in the 15 April 2004 issue of Nature, Michael Shapiro, Melissa Marks, Catherine Peichel, and their colleagues report that a mutation in a noncoding region of the Pitx1 gene accounts for most of the difference in the structure of the pelvic bones of the marine stickleback and its close freshwater cousins.
 The marine sticklebacks
The marine sticklebacks
Here then is a remarkable demonstration of how a single gene mutation can not only be viable but can lead to a major change in phenotype — adaptive evolution. (The changes seem not to have produce true speciation as yet. The marine and freshwater forms can interbreed. In fact, that is how the differences in their hind limbs were found to be primarily due to the expression of Pitx1.)
Sonic hedgehog (Shh), one of the three hedgehog genes found in all vertebrates, is also essential for the development of forelimbs and hindlimbs.
In a remarkable study published in the 20 October 2016 issue of Cell, E. Z. Kvon and colleagues report that snakes have a mutation (a 17-base pair deletion) that inactivates an enhancer of Sonic hedgehog. Transgenic mice whose normal enhancer is replaced by the snake enhancer, fail to form hind legs and have front legs that are badly truncated. But transgenic mice whose own enhancer is replaced by either the human enhancer, or even the enhancer in the lobe-fined coelacanth, develop normally.
Here, then, is another illustration of the crucial role that regulatory regions in the genome have in controlling the expression of genes and thus the phenotype of their owners.
What toolkit proteins do is governed not only by what tissue they are being produced in but also by when they are produced — a phenomenon called heterochrony.
Examples:So the evidence is increasing that what makes the difference between a human and a chimpanzee (or any other pair of animals) is in large measure
The Tc1 mouse is more than simply transgenic —it carries in most of its cells a human chromosome #21. This small chromosome is the one that, when present in a triple dose (trisomy 21), produces Down syndrome in humans. Mice have a similar chromosome that is designated #16.
The question that this remarkable animal could answer: will the genes on human chromosome #21 (105 of them) when present in a mouse nucleus and surrounded by mouse transcription factors and signaling pathways respond
Several transcription factors turn on gene activity in liver cells. As seems to be the case with all transcription factors, the human and mouse versions are close orthologs (95% identical in sequence). Using ChIP analysis, they found that the mouse transcription factors bound to sites along the human chromosome much as the human transcription factors do. (Chromosome #21 does not encode any transcription factors, so all those available in the mouse nucleus were of mouse origin.)
| Tissue | Chromosome | Transcription Factors (TFs) | Sites Bound by TFs | Gene Expression |
| human liver cells | #21 | human TFs | human pattern | human pattern |
| Tc1 liver cells | #21 | mouse TFs | human pattern | human pattern |
| #16 | mouse TFs | mouse pattern | mouse pattern | |
| normal mouse liver cells | #16 | mouse TFs | mouse pattern | mouse pattern |
These workers also examined the pattern of gene expression; that is, the production of messenger RNAs, in the various combinations. They did this using microarrays like those described at this link.
The human genes expressed on chromosome #21 by mouse transcription factors in the Tc1 mouse cells were mostly the same as those turned on by human transcription factors in human cells.
All these lines of evidence point to the following:
Throughout the animal kingdom,So after years of looking for and sequencing open reading frames, the task now will be to analyze the sequence differences — arisen by mutation and evolution — in the intergenic regions that serve as control regions of those genes. An early result of genome analysis: during the radiation of the various mammalian orders, enhancers have diversified (evolved) much more rapidly than have promoters and their associated protein-encoding genes.
| Welcome&Next Search |