One complete set of genes in an organism (a haploid set). [More]
Except for occasional unrepaired damage to its DNA (= mutations), the genome is fixed.
The most common definition: All the messenger RNA (mRNA) molecules transcribed from the genome. [More]
Varies with the differentiated state of the cell and the activity of the transcription factors that turn gene transcription on (and off).
Speaking strictly, one would define the transcriptome as all the RNA molecules — which includes a wide variety of untranslated, nonprotein-encoding RNA [Link to examples] — transcribed from the DNA of the genome. It is now thought that ~75% of our DNA is transcribed into RNA although only 1.5% of this is messenger RNA for protein synthesis.
Two popular definitions:
- All the proteins that can be synthesized by the cell. (The original definition.)
- All the proteins synthesized by a particular cell at a particular time. (The one I shall use in this page.)
All the metabolic machinery, e.g.,
present in a cell at a given time.
Varies with the differentiated state of the cell and its current activities.
The proteome is the protein complement of the genome.
It is quite a bit more complicated than the genome because a single gene can give rise to a number of different proteins through
While we humans probably have only some 21 thousand genes, we probably can make at least 10 times that number of different proteins. The great majority of our genes produce pre-mRNAs that are alternatively-spliced.
The study of proteomics is important because proteins are responsible for both the structure and the functions of all living things. Genes are simply the instructions for making proteins. It is proteins that make life.
At any one time, a human cell might contain some 10,000 different proteins; some (e.g. ribosomal proteins) in great abundance, others (e.g. transcription factors) in lower numbers.
The set of proteins within a cell varies
- from one differentiated cell type to another (e.g. red blood cell vs lymphocyte) and
- from moment to moment, depending on the activities of the cell, e.g.,
- Isolate a homogeneous population of cells (e.g., yeast cells that have just been switched from glucose to galactose as their energy source).
- Extract the contents of the cells and separate the mix of proteins from other components.
- Separate the proteins in the mix by two-dimensional (2D) gel electrophoresis. This separates the proteins
- in one dimension by their electrical charge;
- in the second dimension by their size.
(The procedure is analogous to that used in paper chromatography. [Link])
- Stain the gel to visualize the various spots of protein.
- Punch out a spot.
- Add a protease (e.g., trypsin) to digest the protein in that spot into a mix of peptides.
- Run the mix through a mass spectrometer, which will separate the peptides into sharply-defined peaks.
- Run the resulting data through a database of all known proteins (that have been digested with the same enzyme) to see if you can find a match.
What if there is no match; that is, you have stumbled on an unknown protein?
- Isolate individual peptides from your mix and run one through a mass spectrometer that has been modified to
- first randomly break the peptide into a mix of fragments containing one, two, etc. amino acids
- then measure the mass of each fragment.
- Enter the resulting data into a database that matches the mass data with known pairs, triplets, etc. of amino acids.
- With the aid of overlaps, assemble the fragments to reveal the entire sequence of the peptide.
- "Back-translate" the amino acid sequence to determine what sequence of nucleotides in DNA could encode that peptide.
- Search the genome database for an open reading frame (ORF) that contains that sequence.
- Translate that ORF to get the entire amino acid sequence of your protein.
Some proteins act alone, and the function of many of these has been know for years.
But probably the majority of the proteins in a cell act in concert with others.
Examples:
For this procedure,
- Attach the protein whose partners you wish to find to a solid matrix in a glass column.
- Run a solution containing a mix of possible partners through the column.
- Those that can bind to the target will stick; the others will flow through.
- Pass a buffer through the column which will weaken the binding interactions.
- The partners will wash out and can be identified.
The budding yeast, Saccharomyces cerevisiae, provides an excellent tool for discovering protein partners.
- It can easily be transformed with plasmids containing foreign DNA sequences.
- It can live in either the haploid or diploid condition.
- Haploid cells can fuse to form diploid cells if they are of opposite mating types (designated a and α).
The two-hybrid system also takes advantage of the fact that transcription factors (proteins) usually contain
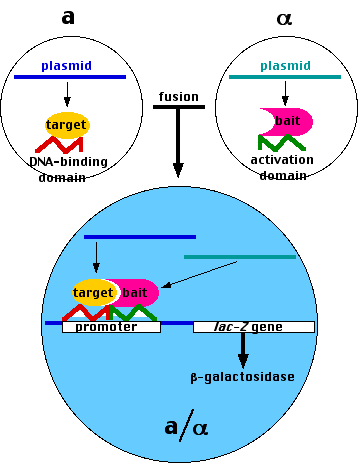
- a DNA-binding domain: a region that binds to a specific sequence of DNA in the promoter of the gene they turn on;
- an activation domain: a region that is needed to activate the assembly of the other components of the transcription apparatus.
- Using recombinant DNA methods, create a plasmid containing
- the DNA encoding the DNA-binding domain of a transcription factor needed to turn on expression of a "reporter gene" such as the lacZ gene that encodes the enzyme β-galactosidase coupled to
- the DNA encoding the "target" protein; that is, the protein whose possible partners you wish to identify.
Insert the plasmid into living haploid yeast of one mating type (e.g., a)
- Using the same methods, create many different plasmids each containing
- the DNA encoding the activation domain of the transcription factor;
- the DNA encoding a possible partner ("bait") protein. (With the help of automated equipment, you can even make plasmids representing each of the entire ~6,000-gene genome of yeast.)
- Insert each of these plasmids into α yeast cells and grow them as separate clones.
- Mate each α clone with the target clone (a).
- If the fusion protein produced by the transcription and translation of a "bait"-containing plasmid
can bind to the fusion protein containing the target,
- the two domains of the transcription factor can interact to turn on expression of the reporter gene (lacZ in our case).
- Grown on an indicator substrate, these colonies will turn blue. [Another example]
- The DNA in these colonies can then be isolated and sequenced.
- The result: identification of the proteins that can associate with the target protein.
Using the two-hybrid method, it has been possible to identify many sets of interacting proteins in yeast and other organisms. (The 23 September 2005 issue of Cell reports the identification of over 3000 interactions among pairs of human proteins.
This method exploits:
- a DNA bacteriophage that infects E. coli;
- its ability to remain infectious even if one of its coat proteins contains segments of a foreign protein.
The method:
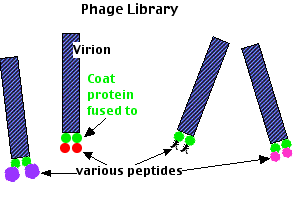
- Transform bacteriophages with a
- random mix of DNA from the organism you are interested in coupled to
- the DNA encoding one of the viral coat proteins.
- Infect E. coli with these phages.
- As the viruses replicate, they will not only propagate the recombinant gene but also express it as a coat protein.
- Both with be incorporated into new virions.
- Harvest the mix of viruses.
- Pass the mixture through an affinity chromatography column to which your "target" protein has been fixed.
- Those viruses that display a piece of foreign protein (peptide) that can bind to the target will stick to it.
- Elute the bound phage with a buffer.
- Repeat steps 6–8 to further enrich your binders.
- Infect E. coli.
- Grow separate colonies (clones).
- Sequence the coat protein gene to find the sequence of the foreign DNA inserted in it.
- Using the codon table, determine the amino acid sequence of the peptide.
- Search databases for a protein containing this sequence.
- Result: another protein that associates with your target protein.
Phage display is also used to make monoclonal antibodies (without the need for mice).
 Protein chips work on much the same principle as DNA chips [Link].
Protein chips work on much the same principle as DNA chips [Link].
- A library of hundreds or even thousands of different proteins from your organism are spotted individually in a known location on a chip.
- The chip is flooded with a solution of the protein whose partners you seek.
- Any proteins on the chip that are potential binding partners will bind your test protein.
- Adding a fluorescent "tag" permits these to be identified.
Although simple in principle, protein chips are far more difficult to work with than DNA chips because proteins
- vary enormously in their chemistry (e.g., hydrophobic vs hydrophilic);
- bind to each other by several types of noncovalent interactions. [Link]
Fragments of DNA, in contrast, vary only in their nucleotide sequence and all bind their partners by simple Watson-Crick base pairing.
The clearest picture of how different proteins interact with one another to form functional complexes will come from determining the 3D structure of the complex.
There are two methods:
- x-ray crystallography;
- nuclear magnetic resonance (NMR) spectroscopy.
X-ray crystallography requires that you be able to crystallize the protein. This is often a difficult task and especially difficult for complexes of two or more proteins.
Here are some links to 3D images of proteins.
Note that although in both cases the proteins are binding to DNA, they are also binding to each other (as homodimers).
NMR spectroscopy has been especially useful in producing 3D images of proteins that cannot be crystallized.
30 October 2022



 Protein chips work on much the same principle as DNA chips [Link].
Protein chips work on much the same principle as DNA chips [Link].