Humans are members of the order of placental mammals known as primates.
The earliest primates were probably arboreal (tree-living) and resembled today's lemurs and tarsier.Associated with these adaptations was an increase in the relative size of the brain, particularly evident in the motor, visual, and auditory regions of the cerebral cortex.
Descendants of these early primates include the Over the next 20 million years or so, the gibbons ("lesser apes") and 3 groups of "great apes"
Over the next 20 million years or so, the gibbons ("lesser apes") and 3 groups of "great apes"

A phylogenetic tree of living hominoids based on DNA-DNA hybridization data (left — the work of Charles Sibley and Jon Ahlquist at Yale University) supports the picture above.
The numbers represent the differences between various measured T50H values split up and apportioned to each branch. [Link to a description of the DNA-DNA hybridization method and the meaning of T50H.]
For example, the T50H difference between the common chimpanzee (Pan troglodytes) and the pygmy chimpanzee or bonobo (Pan paniscus) is 0.73. Assuming that their DNA has evolved at the same rate since they diverged, each branch is given one-half that value.
The difference between the T50H values of humans and the chimpanzees is about 1.6, whereas that between the chimpanzees and the gorilla is about 2.3. This indicates that humans and chimpanzees have shared a common ancestor more recently than chimpanzees and gorillas have.
The time scale was calibrated using fossil evidence that the line to the orangutan diverged some 13–16 million years ago.
| These results as well as actual genome sequences [Link to The Human and Chimpanzee Genomes] have led to an even greater appreciation of the close evolutionary relationship of humans to chimpanzees. |
All of the earliest hominin remains have been found in Africa. Specimens of a species called Australopithecus afarensis appear in deposits dating about 3.3 million years ago. An almost complete skeleton of this species is a young female dubbed "Lucy". Lucy differed from her primate forebears by always walking upright. We know this because of the way in which her legs were attached to her hip girdle and the fact that her backbone was attached to the base of the skull rather than at its back as it is in four-legged mammals. To clinch the matter, fossil footprints have been found that show that A. afarensis walked upright.
 |
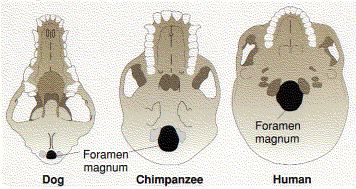
| The opening in the skull where the spinal cord enters is called the foramen magnum. It is at the rear of the skull of quadrupedal mammals like the dog, shifted forward in knuckle-walking hominines like the chimpanzee, and farther forward still in bipedal hominins like Lucy (Australopithecus afarensis) and modern humans. |
Over the next 2 million years, several independent hominin groups evolved. All of these forms were more or less contemporaries and, probably, sympatric – that is, lived in the same regions of Africa. These included:
Remains of Homo habilis have been found along with primitive stone implements resembling spearheads and the like. This indicates that Homo habilis made and used tools. Perhaps the demands of toolmaking and a way of life based on hunting for the benefit of the community created a selection pressure for a rapid increase in the size of the brain. In any case, Homo habilis had a brain volume of approximately 600 ml, one-third again as large as that of Australopithecus.
Although Homo erectus arose in Africa (where it is often called Homo ergaster), it soon spread into Europe and throughout Asia ("Java man" and "Peking [Beijing] man").
Over the past 8 years a group of 5 hominin fossils have been excavated and analyzed from a single site in Dmanisi in the nation of Georgia (at the east end of the Black Sea). All five of these skeletal remains, including skulls, appear to have been fossilized about 1.8 million years ago but within a few hundred years of each other. So they surely represent individuals of a single species.
What makes them so remarkable is that they differ substantially from one another in a range of traits, such as skull morphology, that overlap with those of fossils attributed to Homo erectus and also Homo habilis from various parts of Africa and Eurasia. In other words, perhaps the various fossils assigned to Home habilis and Homo erectus/ergaster are actually all members of a single species showing a range of variation no greater than is found today within the human population. In fact, estimates of their body size overlap the low end of today's humans although their brains are much smaller (546–730 ml compared with our 1,100–1,400 ml).
If these five fossils had been found in different parts of Africa, paleontological "splitters" would have given them different species names. But found together in Dmanisi, they provide strong support for "lumpers".
The fossils at Dmanisi appear to have solved some of these problems. It now seems likely that fossils attributed in the past to Homo ergaster, Homo habilis, Homo rudolfensis (all in Africa), Pithecanthropus erectus (in Java), etc., are all members of single evolving species, Home erectus, showing variations with the passage of time (over a million years) and over large geographical distances.
 |
| Approximate span of time during which various hominin groups appear in the fossil record. The specimens found in Dmanisi suggest that Homo habilis and Homo erectus are actually members of the same species (to be called Homo erectus). |
Not until some 200,000 years ago do clearly modern humans, Homo sapiens, appear in the fossil record. Where did they come from? Two quite different scenarios have been proposed.
1. One scenario ("Recent African Origin") is that modern humans arose (like all the other hominins) in Africa. Some 60,000 years ago, they began to spread into Europe and Asia where, despite some interbreeding, they eventually replaced the preexisting populations of Neanderthals (as well as a related group, the Denisovans). Today we still retain some Neanderthal DNA and, in some populations, some Denisovan DNA. The DNA testing company 23andMe tells me that almost 4% of my DNA is derived from the Neanderthals!)
2. A second scenario ("multiregional evolution") claims that modern humans slowly evolved throughout Eurasia from preexisting interbreeding populations of primitive H. sapiens, Neanderthals, and Denisovans (all descendants of H. erectus).
Computer analysis of the molecules (proteins or DNA) of contemporary species can be used to generate a genealogical or phylogenetic tree linking them. [Link to discussion]
To do a similar analysis for humans requires finding a molecule that has mutated more rapidly than molecules like cytochrome c or the rRNAs that have helped reveal more ancient relationships.
A noncoding portion of the mitochondrial DNA (mtDNA) molecule was chosen on the assumption that it would not have been so constrained by the pressures of natural selection. Mutations accumulate more freely in noncoding DNA.
The choice of mitochondrial DNA had two other advantages:Over 600 base pairs of a noncoding region of mtDNA were sequenced in specimens collected from over 200 people. The subjects represented populations from all parts of the world, including a number of different tribal groups in Africa.
Over 100 different sequences were found. In almost all cases, two or more people sharing the same sequence also belonged to the same population. The greatest diversity of sequences was found among the various African populations.
Computer analysis produced many possible family trees by which the present-day sequences could be derived ultimately from a single ancestral sequence. Because of the maternal inheritance of mtDNA, this work was interpreted as showing that all present mtDNA is derived from a single ancestral mtDNA belonging to a woman who lived in Africa some 160,000 years ago. She was nicknamed "Mitochondrial Eve", the mother of us all.
Some of the implications from these data are more dramatic than is justified. Mitochondrial Eve was only one of many mothers in the population living then. The story of Ariaantje and Gerrit Jansz will help explain how her mitochondrial DNA ended up being the only one represented in today's human population.
Some 30,000 people living in South Africa today carry the gene for porphyria that either Ariaantje or Gerrit brought with them from Holland in the 1680s. Every one of these people can find a genealogical line leading back to this couple. But that does not mean that Ariaantje or Gerrit by themselves founded this group. Although each of the 30,000 people with the porphyria gene can trace a genealogical line back to this couple, they could do the same for many couples living at that time. We each had 4 grandparents, 8 great-grandparents, 16 great-great-grandparents, and so on. Because of the restricted maternal inheritance of mtDNA, every time a couple descended from the population to which Eve belonged failed to have any daughters, another line of mtDNA would have died out. By chance alone, Eve's mtDNA eventually was the only one left. This outcome is similar to the operation of genetic drift in neutral alleles of nuclear genes: in due course a given allele reaches a gene frequency of 100% or disappears entirely [Link to discussion].

In principle, the accumulation of mutations in a gene during the course of evolution could provide a clock to measure the passage of time. The clock must tick at a constant rate, and one must be able to determine the rate; that is, to calibrate the clock. For some genes, the accumulation of mutations does appear to occur at a steady rate. Consider the sequence differences that are found between 3 contemporary species in figure: A, B, and C.
On other grounds (e.g. fossil evidence) we know that A and B diverged after they shared a common ancestor with C. If the total number of sequence differences between A and C is the same as that between B and C, then the rate of mutation in the lines leading to A and B has been the same since they diverged. This is because we know that A and B have been evolving for the same length of time since they diverged from their common ancestor with C. Even though A and B might have been subjected to different pressures of natural selection, the clock has ticked steadily. This type of analysis has shown that many (but not all) genes accumulate mutations at a steady rate.
So the clock may be steady, but how fast is it ticking? To determine this requires an independent way of dating the time of divergence of two genealogical lines. If you know that the node from which C, A, and B branched represents an ancestor whose fossils have been dated as, say, 30 million years old, and half the genetic difference between A and B (10) is one-half of that between them and this ancestor (20), then A and B diverged from each other 15 million years ago.
Using this approach, the split between the line leading to chimpanzees and that to humans has been dated at 5–7 million years ago. Then comparing the mtDNA of chimpanzees with the much smaller differences in the mtDNAs of all modern humans places our common ancestor ("Eve") at only 200,000 or so years ago.
But if all modern humans are descended from a population that lived in Africa just 200,000 years ago, then as they emigrated out of Africa and spread throughout Europe and Asia, they must have replaced the various H. erectus descendant populations (including Neanderthals and Denisovans) that had preceded them some 1.8 million years before.
Neanderthals, for example, appear in the fossil record in Europe and Asia long before the appearance of modern H. sapiens and coexisted with H. sapiens until disappearing some 40 thousand years ago. As we saw above, there is now genetic evidence of hybridization between the two groups. This would make Neanderthals a subspecies of H. sapiens. In any case, before we can be certain of our origins, we need more data — fossils as well as molecules.
One thing is clear. Despite the differences that exist today between the various racial groups, humans actually represent a remarkably homogeneous population. (Genetic differences among the members of a single species of chimpanzee are five times greater than those among humans.) Furthermore, whatever genetic differences did arise in the separate gene pools in Africa, Europe, Asia, and so on, these now appear to be diminishing. Gene flow between these gene pools will probably accelerate as geographical barriers break down and, some time later, cultural barriers to gene flow fall as well.
The outlook for the future is for an accelerating process of gene flow between the various human gene pools, leading ultimately to a worldwide, randomly-breeding ("panmictic") population. If and as this prediction is fulfilled, we may hope that the many social barriers that have set groups of humans in conflict with one another will disappear as well.
| Welcome&Next Search |