Cells must be ready to respond to essential signals in their environment. These are often chemicals in the extracellular fluid (ECF) from:
- distant locations in a multicellular organism — endocrine signaling by hormones;
- nearby cells — paracrine stimulation by cytokines;
- or even secreted by themselves ( = autocrine stimulation).
They may also respond to
- molecules on the surface of adjacent cells (e.g. producing contact inhibition);
- molecules (e.g., mRNAs, proteins) that reach them in exosomes secreted by distant cells.
Signaling molecules may trigger:
- an immediate change in the metabolism of the cell (e.g., increased glycogenolysis when a liver cell detects adrenaline);
- an immediate change in the electrical charge across the plasma membrane (e.g., the source of action potentials);
- a change in the gene expression — transcription — within the nucleus. (These responses take more time.)
It is the third category that is the topic of this page.
So this page examines some of the major pathways by which the arrival of a chemical signal at a cell turns on a new pattern of gene expression.
Two categories of signaling molecules (steroids and nitric oxide) diffuse into the cell where they bind internal receptors.
The others, e.g., proteins, bind to receptors displayed at the surface of the cell. These are transmembrane proteins whose
- extracellular portion has the binding site for the signaling molecule (the ligand);
- intracellular portion activates proteins in the cytosol that in different ways eventually regulate gene transcription in the nucleus.
Steroids are small hydrophobic molecules that can freely diffuse across the plasma membrane, through the cytosol, and into the nucleus.
Receptors
Steroid receptors are homodimers of zinc-finger proteins that reside within the nucleus (except for the glucocorticoid receptor which resides in the cytosol until it binds its ligand).
Until their ligand finds them, some steroid receptors within the nucleus associate with histone deacetylases (HDACs), keeping gene expression repressed in those regions of the chromosome.
Ligands
Some steroids that regulate gene expression:
Mechanism
- The steroid binds its receptor.
- The complex
- NO diffuses freely across cell membranes.
- There are so many other molecules with which it can interact, that it is quickly consumed close to where it is synthesized.
- Thus NO acts in a paracrine or even autocrine fashion — affecting only cells near its point of synthesis.
Receptors
The signaling functions of NO begin with its binding to protein receptors in the cell. The binding sites can be either:
- a metal ion in the protein or
- one of its S atoms (e.g., on cysteine).
Mechanism
In either case, binding triggers an allosteric change in the protein which, in turn, triggers the formation of a "second messenger" within the cell. The most common protein target for NO seems to be guanylyl cyclase, the enzyme that generates the second messenger cyclic GMP (cGMP).
Receptors
These are transmembrane proteins that wind 7 times back and forth through the plasma membrane. Humans have over 800 different GPCRs.
- Their ligand-binding site is exposed outside the surface of the cell.
- Their effector site extends into the cytosol.
Ligands
Some of the many ligands that alter gene expression by binding GPCRs:
- protein and peptide hormones such as:
- Serotonin and GABA (which affect gene expression in addition to their role as neurotransmitters)
Mechanism
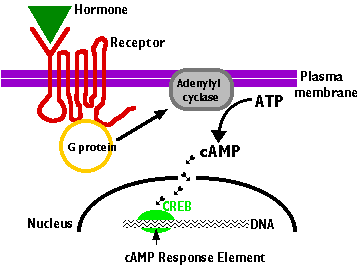
- The ligand binds to a site on the extracellular portion of the receptor.
- Binding of the ligand to the receptor
- activates a G protein associated with the cytoplasmic C-terminal.
- This initiates the production of a "second messenger". The most common of these are
- The second messenger, in turn, initiates a series of intracellular events (shown here as short arrows) such as
- phosphorylation and activation of enzymes
- release of Ca2+ into the cytosol from stores within the endoplasmic reticulum
- In the case of cAMP, these enzymatic changes activate the transcription factor CREB (cAMP response element binding protein)
- Bound to its response element
5' TGACGTCA 3'
in the promoters of genes that are able to respond to the ligand, activated CREB turns on gene transcription.
- The cell begins to produce the appropriate gene products in response to the signal it had received at its surface.
In addition to their roles in affecting gene expression, GPCRs regulate many immediate effects within the cell that do not involve gene expression. Links to some examples.
Turning GPCRs Off
A cell must also be able to stop responding to a signal.
Several mechanisms cooperate in turning GPCRs off.
- When activated, the Gα subunit of the G protein swaps GDP for GTP. However, the Gα subunit is a GTPase and quickly converts GTP back to GDP restoring the inactive state of the receptor. [Link to a discussion.]
- The receptor itself is phosphorylated by a kinase, which not only reduces the ability of the receptor to respond to its ligand but
- recruits a protein, β-arrestin, which
- further desensitizes the receptor, and
- triggers the breakdown of the second messengers of the GPCRs:
- cAMP for some GPCRs
- DAG for others.
Receptors
Frizzled receptors, like GPCRs, are transmembrane proteins that wind 7 times back and forth through the plasma membrane.
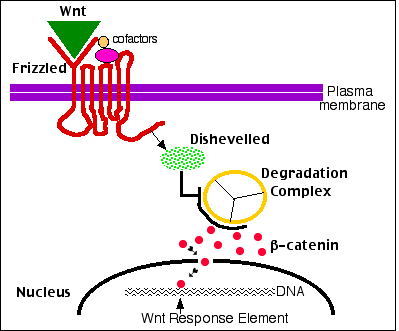
- Their ligand-binding site is exposed outside the surface of the cell.
- Their effector site extends into the cytosol.
Ligands
Their ligands are Wnt proteins. These get their name from two of the first to be discovered, proteins encoded by
- wingless (wg) in Drosophila and its homolog
- Int-1 in mice.
The roles of β-catenin
β-catenin molecules connect actin filaments to the cadherins that make up adherens junctions that bind cells together.
Any excess β-catenin is quickly destroyed by a multiprotein degradation complex. (One component is the protein encoded by the APC tumor suppressor gene.)
The degradation complex
- phosphorylates β-catenin so it can
- have ubiquitin molecules attached to prepare it for destruction in
- proteasomes (not shown).
But undegraded β-catenin takes on a second function: it becomes a potent transcription factor.
Mechanism
- The binding of a Wnt ligand to Frizzled (done with the aid of cofactors) activates Frizzled. This, in turn,
- activates a cytosolic protein called Dishevelled.
- Activated Dishevelled inhibits the β-catenin degradation complex so
- β-catenin escapes destruction by proteasomes and is free to enter the nucleus where
- it binds to the promoters and/or enhancers of its target genes.
(Note the similarities to the strategy used by the NF-κB signaling pathway.)
Wnt-controlled gene expression plays many roles:
- in embryonic development (e.g., a gradient from low at the future head to high at the future tail establishes the anterior-posterior axis throughout the metazoa);
- it guides regeneration
- as well as regulating activities in the adult body.
Receptor
Patched (PTCH) — a 12-pass transmembrane protein embedded in the plasma membrane.
Ligands
Secreted hedgehog proteins (HH) that diffuse to their targets. Vertebrates have three hedgehog genes encoding three different proteins. However, hedgehog was first identified in Drosophila, and the bristly phenotype produced by mutations in the gene gave rise to the name.
Mechanism
- When there is no hedgehog protein present, the patched receptors bind a second transmembrane protein called smoothened (SMO).
- However, when HH protein binds to patched, the SMO protein separates from PTCH
- enabling SMO to activate a zinc-finger transcription factor designated GLI.
- GLI migrates into the nucleus when it activates a variety of target genes.
Hedgehog signaling plays many important developmental roles in the animal kingdom. For example,
- wing development in Drosophila;
- development of limbs in vertebrates. Snakes, which have no limbs, are descended from limbed ancestors. However, during their evolution they acquired a mutation in an enhancer that normally drives hedgehog signaling but has been inactivated in them.
Mutations or other sorts of regulatory errors in the hedgehog pathway are associated with a number of birth defects as well as some cancers. Basal-cell carcinoma, the most common skin cancer (and, in fact, the most common of all cancers in much of the world), usually has mutations causing - extra-high hedgehog or
- suppressed patched activity (both leading to elevated GLI activity).
This pathway is found throughout the animal kingdom. It differs from many of the other signaling pathways discussed here in that the ligands as well as their receptors are transmembrane proteins embedded in the plasma membrane of cells. Thus, signaling in this pathway requires direct cell-to-cell contact.
Receptors
Notch proteins are single-pass transmembrane glycoproteins. They are encoded by four genes in vertebrates. However, the first notch gene was discovered in Drosophila where its mutation produced notches in the wings.
Ligands
Their ligands are also single-pass transmembrane proteins. There are many of them and often several versions within a family (such as the serrate and delta protein families).
Mechanism
When a cell bearing the ligand comes in contact with a cell displaying the notch receptor, the external portion of notch is cleaved away from the cell surface (and engulfed by the ligand-bearing cell by endocytosis). The internal portion of the notch receptor is cut away from the interior of the plasma membrane and travels into the nucleus where it activates transcription factors that turn the appropriate genes on (and off).
It would appear that proper development of virtually all organs (e.g., brain, immune system, pancreas, GI tract, heart, blood vessels, mammary glands) depends on notch signaling. Notch signaling appears to be a mechanism by which one cell tells an adjacent cell which path of differentiation to take (or not take).
Defects in notch signaling have been implicated in some cancers, e.g. melanoma.
Dozens of cytokine receptors have been discovered. Most of these fall into one or the other of two major families:
- Receptor Tyrosine Kinases (RTKs) and
- Receptors that trigger a JAK-STAT pathway.
Receptors
The receptors are transmembrane proteins that span the plasma membrane just once. 58 different RTKs are found in humans.
Ligands
Some ligands that trigger RTKs:
Mechanism
- Binding of the ligand to two adjacent receptors forms an active homodimer.
- This activated dimer is a tyrosine kinase; an enzyme that attaches phosphate groups to certain tyrosine (Tyr) residues — first on itself, then on other proteins converting them into an active state.
- Many of these other proteins are also tyrosine kinases (the human genome encodes 90 different tyrosine kinases) and in this way a cascade of expanding phosphorylations occurs within the cytosol.
Turning RTKs Off
A cell must also be able to stop responding to a signal. For growth factor receptors, failure to do so could lead to uncontrolled mitosis = cancer.
For the RTKs, this is done by quickly engulfing and destroying the ligand-receptor complex by receptor-mediated endocytosis.
Proto-Oncogenes
It should not be surprising that anything which leads to the inappropriate expression of receptors that trigger cell division could lead to cancer (uncontrolled cell division).
An example:
The gene (EGFR) encoding the receptor for epidermal growth factor (EGF) is a proto-oncogene. Mutations in EGFR are common in several human cancers. Cetuximab (Erbitux®), a monoclonal antibody that blocks the epidermal growth factor receptor, shows promise against some colorectal cancers as well as cancers of the head and neck.
Two tyrosine kinase inhibitors
- gefitinib (Iressa®) and
- erlotinib (Tarceva®)
block the action of the EGF receptors on the cells of certain lung cancers and have shown some promise against these cancers.
Mutant versions of some of the "second-order" kinases are also associated with cancer:
- The oncogene SRC encodes a mutated version of a normal tyrosine kinase associated with the inner face of the plasma membrane.
- The fusion protein BCR/ABL produced by the Philadelphia chromosome activates constitutively (all the time) the cytosolic tyrosine kinase ABL that normally would be activated only when the cell is stimulated by a growth factor (e.g., PDGF).
The result: chronic myelogenous leukemia (CML).
A promising treatment: Imatinib mesylate (Gleevec® also known STI571). This molecule fits into the active site of the ABL protein preventing ATP from binding there. Without ATP as a phosphate donor, the ABL protein cannot phosphorylate its substrate(s).
- RAF. This kinase participates in a signaling pathway that links RTKs to gene activation. Binding of a ligand to the RTK activates an intracellular molecule called RAS, which then activates RAF. In mammals, this pathway promotes mitosis. Excessive activity of the RAS gene or mutations in RAS and/or RAF are associated with many types of cancer so RAS and RAF are proto-oncogenes.
Receptors
These consist of 2 identical single-pass transmembrane proteins (i.e., homodimers) embedded in the plasma membrane. Each of their cytoplasmic ends binds a molecule of a Janus kinase ("JAK").
Ligands
Many ligands trigger JAK-STAT pathways:
Mechanism
- Binding of the ligand activates the JAK molecules which
- phosphorylate certain tyrosine (Tyr) residues on each other as well as on one or another of several STAT ("Signal Transducer and Activator of Transcription) proteins.
- These, in turn, form dimers which
- enter the nucleus and
- bind to specific DNA sequences in the promoters of genes that
- begin transcription.
The JAK-STAT pathways are much shorter and simpler than the pathways triggered by RTKs, and so the response of cells to these ligands tends to be much more rapid.
Receptors
Two types of single-pass transmembrane proteins that, when they bind their ligand, become kinases that attach phosphate groups to serine and/or threonine residues of their target proteins.
Ligands
Ligands for these receptors include:
Mechanism
- The ligand binds to the extracellular portion of the receptors,
- which then phosphorylate one or more
SMAD proteins in the cytosol.
- The SMAD proteins move into the nucleus where they
- form heterodimers with another SMAD protein designated SMAD4.
- These dimers bind to a DNA sequence (CAGAC) in the promoters of target genes and — with the aid of other transcription factors —
- enhance, or repress, as the case may be,
- gene transcription.
Tumor-Suppressor Genes
The TGF-β signaling pathway suppresses the cell cycle in several ways. So it is not surprising that defects in the pathway are associated with cancer.
Mutations in the genes encoding
- the TGF-β receptors as well as
- of the SMAD proteins
are found in many cancers including pancreatic and colon cancer.
Thus all these genes qualify as tumor-suppressor genes.
TNF-α is made by macrophages and other cells of the immune system.
Receptors
Trimers of 3 identical cell-surface transmembrane proteins.
Ligands
- TNF-α (hence the name)
- Lymphotoxin (LT; also known as TNF-β)
Mechanism
- NF-κB resides in the cytosol bound to an inhibitor called IκB.
- Binding of ligand to the receptor triggers phosphorylation of IκB
- IκB then becomes ubiquinated and destroyed by proteasomes.
- This liberates NF-κB so that it is now free to move into the nucleus where
- it acts as a transcription factor binding to the promoters and/or enhancers of more than 60 genes:
- NF-κB got its name from its discovery as a transcription factor bound to the enhancer of the kappa light chain antibody gene.
- However, it also turns on the genes encoding IL-1 and other cytokines that promote inflammation.
| The immunosuppressive and anti-inflammatory effects of glucocorticoids are caused by their enhancing the production of IκB. |
- NF-κB also turns on genes needed for cell proliferation, cell adhesion, and angiogenesis.
In May 2003, the US FDA approved a proteasome inhibitor, called bortezomib (Velcade®) for treatment of multiple myeloma, a cancer of plasma cells. [More]
The monoclonal antibody infliximab binds to TNF-α, and shows promise against some inflammatory diseases such as rheumatoid arthritis.
|
| In addition to its effects of gene expression, activation of the TNF-α receptor can lead to apoptosis of the cell. [Link] |
T cells use a transmembrane dimeric protein as a receptor for a particular combination of an antigen fragment nestled in the cleft [View] of a histocompatibility molecule.
Receptors
Ligands
Mechanism
Activation of the TCR (when aided by costimulator molecules also present in the plasma membrane — View)
- causes a rise in intracellular Ca2+ which
- activates calcineurin, a phosphatase which
- removes phosphate from NF-AT ("Nuclear Factor of Activated T cells").
- Dephosphorylated NF-AT enters the nucleus, and with the help of accessory transcription factors (designated AP-1), binds to the promoters of some 100 genes expressed in activated T cells.
The immunosuppressant drugs tacrolimus and cyclosporine inhibit calcineurin thus reducing the threat of transplant rejection by T cells.
Activation of the TCR, when accompanied by an as-yet-unidentified second signal, causes NF-AT to associate with a different transcription factor (designated Foxp3). Instead of activating the T cell, this turns on genes that convert the cell into a suppressive regulatory T cell (Treg) instead — Link.
- The signaling systems discussed here are only some of the major players,
- and even the description of these is greatly oversimplified because
- many of these signaling pathways interconnect with one another (creating "cross talk").
- While cross talk might seem to create problems (for our comprehension as well as for the cell!), in fact
- it is probably essential to precisely modulate the genetic response to a variety of ligands reaching the cell at the same time and in varying intensities.
Reading this page may help explain why such a large proportion of the genome of animals is devoted to genes involved in cell signaling.
27 September 2019

