
φX174 (phiX174) is a virus that infects the bacterium E. coli. Hence φX174 is a bacteriophage.
Each complete infectious particle (virion) of φX174 consists of a protein coat which envelopes a core that contains both protein and DNA. The coat of the virus contains 60 molecules each of two proteins (F and G) and 12 molecules of another protein (H). The core of the virion contains one molecule of DNA and 60 copies of a fourth protein, the J protein. The DNA molecule is single-stranded (ssDNA) and is in the form of a closed circle. It contains 5386 nucleotides. This tiny genome was the first DNA genome ever to be sequenced (by Fred Sanger in 1976).
When φX174 attaches to its host, its ssDNA molecule is inserted into the cell. Here the DNA strand (+) serves as the template for the synthesis of a complementary (−) strand. The two strands form a double helix [view below] which then replicates itself several times.
The minus strands of these DNA molecules then serve as templates for the synthesis of
| Protein | Number of Amino Acids |
| A | 513 |
| A* | 341 |
| B | 120 |
| C | 86 |
| D | 152 |
| E | 91 |
| F | 427 |
| G | 175 |
| H | 328 |
| J | 38 |
| K | 56 |
| Total | 1986 |
The protein-synthesizing machinery of the host cell translates the viral mRNA molecules into 11 different kinds of proteins. Four of these are the four (F, G, H, and J) that will be incorporated into new virions. As for the other 7 proteins
The 11 proteins encoded by φX174 DNA range in size from the A protein, which contains 513 amino acids, to the J protein, which contains only 38. The 11 proteins together contain a total of 1986 amino acids (the A* protein is simply a shortened version of the A protein). This raises a question. With 3 nucleotides needed to specify one amino acid, φX174 would need 5958 nucleotides to encode 1986 amino acids (5958/3 = 1986). But its DNA molecule contains 5386 nucleotides, only enough to encode 1795 amino acids. Furthermore, it turns out that 217 of the nucleotides do not encode anything, although some of them provide control signals. So there are only 5169 coding nucleotides, and we would expect them to be able to encode only 1723 amino acids. How does φX174 dictate the assembly of the remaining amino acids?
It does so by using some stretches of nucleotides to encode two different sequences of amino acids. The principle is really quite simple. It involves reading the codons it two different "reading frames", that is, grouping the nucleotides in shifted clusters of three.
For example, the sequenceLink to a table given the DNA codons for all the amino acids.
φX174 actually uses two of these and, as you can see, each encodes a totally different sequence of amino acids.
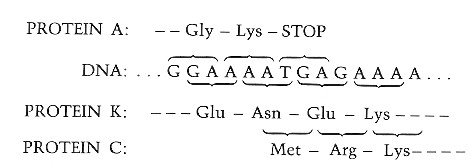
There is even one spot where a single nucleotide (A) participates in three different codons:
Why overlapping genes? φX174 is one of the tiniest viruses. Its use of overlapping genes enables it to increase the amount of information it can store in a given amount of DNA.
Not only was the φX174 genome the first to be sequenced, it was also the first to be chemically synthesized in the laboratory. When introduced into E. coli, this synthetic molecule was fully infectious able to produce intact viruses.
 On the right is an electron micrograph of the double-stranded φX174 DNA extracted from infected E. coli cells. The bar represents 0.5 µm. (Courtesy of David T. Denhardt.)
On the right is an electron micrograph of the double-stranded φX174 DNA extracted from infected E. coli cells. The bar represents 0.5 µm. (Courtesy of David T. Denhardt.)
| Welcome&Next Search |