| Index to this page |
T2 and its close relative T4 are viruses that infect the bacterium E. coli. The infection ends with destruction (lysis) of the bacterial cell so these viruses are examples of bacteriophages ("bacteria eaters").
They have been enormously useful in genetic studies because:Another page — Bacteriophage Genetics — describes how T2 is used to map the order and relative spacing of genes on the single circular molecule of DNA that is the virus's genome.
Here let us see how T4 can be used to:In Bacteriophage Genetics, we examined mutation of a gene designated r, for "rapid lysis" [Link].
It turned out that actually there are three different gene loci — rI, rII, and rIII — mutations in any one of which produced a rapid-lysis phenotype. But, in addition, there were many mutations found in each of these. Could wild-type virus be formed by recombination between mutations within the same gene? Seymour Benzer decided to find out.As we saw in Bacteriophage Genetics, the recombination frequency between different genes is low (on the order of 10-2).
One would expect that recombination frequencies between mutations in a single gene would be far lower (10-4 or less).
Fortunately Benzer could exploit a phenomenon to enable him to detect such rare events: rII mutants as well as wild-type T4 can infect and complete their life cycle in a strain of E. coli designated B. However, while rII mutants can infect a strain of E. coli designated K, they cannot complete their life cycle in strain K. Wild-type T4 can.
The procedure was to infect strain B in liquid culture with two mutants to be tested (designated here as rx and ry).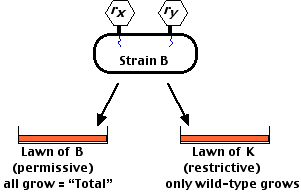 After incubation, these were plated on a lawn of:
After incubation, these were plated on a lawn of:
Recombination Frequency = 2 × number of wild-type plaques (strain K plaques) ÷ total number of plaques (on strain B).
You have to double the number found on strain K because you only see one-half the recombinants — the other half consists of double mutants. Using this technique, Benzer eventually found some 2000 different mutations in the rII gene. The recombination frequency between some pairs of these was as low as 0.02.In other words,
The relative order and spacing of any two point mutations in a single gene like rII can be done using the procedure describe in Bacteriophage Genetics. But with some 2000 different mutations to test, the process would be tremendously time-consuming. (Even using the procedure to be described now, Benzer spent some 10 years on the project.)
Benzer was able to speed up the mapping process by taking advantage of the discovery that some of his mutants did not have point mutations but deletions instead. In contrast to the properties of T4 viruses with point mutations, T4 viruses with deletions in rIIDeletions can be mapped by the same procedure used for point mutations. Simply cross pairs of deletion mutants and see if they produce progeny that can grow on E. coli strain K.
Here is a hypothetical example. Each of 6 strains of deletion mutants are crossed with each of the others.| Strains | 1 | 2 | 3 | 4 | 5 | 6 | |
| 1 | 0 | 0 | + | 0 | 0 | 0 | 1 and 3 do not overlap |
| 2 | 0 | + | + | 0 | 0 | must shift 4 away from 2 | |
| 3 | 0 | + | + | 0 | 6 must extend under 3 | ||
| 4 | 0 | + | + | right-hand end of 4 must be removed from over 6 | |||
| 5 | 0 | + | left-hand end of 6 must not overlap 5 but must continue to overlap 2. ∴ shorten right-hand end of 5 |
||||
| 6 | 0 |
From the results, one can draw a map showing the order and relative size of the deletions.
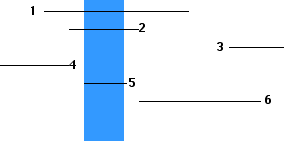
With such a deletion map, one can now quickly map the location of point mutations by coinfecting each of the different deletion strains (here 1–6) with the mutant strain ("x").
There is no longer any need to count plaques; simple see whether there is growth or not.
| Coinfect with strain | 1 | 2 | 3 | 4 | 5 | 6 |
| and mutant "x" Results → |
0 | 0 | + | + | 0 | + |
From these results, we learn that the point mutation "x" is located on the T4 DNA within the region shown above in blue.
As we saw above, rapid lysis (r) mutants were found that mapped to three different regions of the T4 genome: rI, rII, and rIII.
This meant that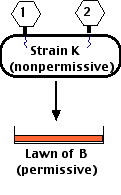
| Mutant strains | 1 | 2 | 3 | 4 | 5 |
| 1 | 0 | 0 | + | 0 | + |
| 2 | 0 | + | 0 | + | |
| 3 | 0 | + | 0 | ||
| 4 | 0 | + | |||
| 5 | 0 |
From these results, you can deduce that these 5 rII mutants fall into two different complementation groups, which Benzer designated
Benzer coined the term cistron for these genetic units of function. But today, we simply modify earlier concepts of the "gene" to fit this operational definition.
| Try some problems on deletion mapping and complementation. |
| Welcome&Next Search |